I joined the Song Lab at Wake Forest University School of Medicine as a high school intern and was fortunate to collaborate with Professor Song on two research projects. Excited by the application of neural networks in medicine and inspired by Prof. Song’s previous work, I initiated two studies detailed below. I spent, on average, an hour every day during the school year and 8 hours a day during summer on research. Prof. Song primarily specializes in the field of medicine; therefore, I undertook the computer science aspects of my research independently.
PINet: Privileged Information Improve the Interpretability and generalization of structural MRI in Alzheimer’s Disease
Introduction
PINet is a neural network model devised to detect Alzheimer’s disease from MRI images even before symptoms occur. The research idea focusing on early Alzheimer’s detection originated from my grandpa’s diagnosis of late-stage Alzheimer’s. Before his diagnosis, my parents and I had attributed his occasional forgetfulness and throwing tantrums to being old and angry over our lack of company, only to realize his condition when it was already too late for any form of intervention. We witnessed his condition develop into dementia and his memory and life were slowly taken away from him. I devoted time to researching the early diagnosis and intervention of Alzheimer’s and realized the potential for neural networks to play a pivotal role.
This paper was accepted (acceptance rate 29%) by the ACM-BCB conference as a rapid-fire paper (top 25% of accepted papers)
Abstract
The irreversible and progressive atrophy of Alzheimer’s Disease resulted in a continuous decline in thinking and behavioral skills. To date, CNN classifiers have been widely applied to assist in the early diagnosis of AD and its associated abnormal structures. However, most existing black-box CNN classifiers relied heavily on limited MRI scans and used little domain knowledge from previous clinical findings. In this study, we proposed a framework, PINet, to consider previous domain knowledge as Privileged Information (PI), and peak into the black-box in the prediction process. PINet uses a Transformer-like fusion module Privileged Information Fusion (PIF) to iteratively calculate the correlation of the features between image features and PI features, and project the features into a latent space for classification. PINet is suitable for neuro-imaging tasks and we demonstrated its application in Alzheimer’s Disease using structural MRI scans from the ADNI dataset. The F1-score showed that PINet is more robust in transferring to a new dataset, with an approximately 2% drop (from 0.9471 to 0.9231), while the baseline CNN methods had a 29% drop (from 0.8679 to 0.6154). Our best model was trained under the guidance of 12 selected ROIs, majoring in the structures of the Temporal Lobe and Occipital Lobe. In summary, PINet incorporates domain knowledge as PI to train the CNN model, and the selected PI introduces both interpretability and generalization ability to the black box CNN classifiers.
Overview
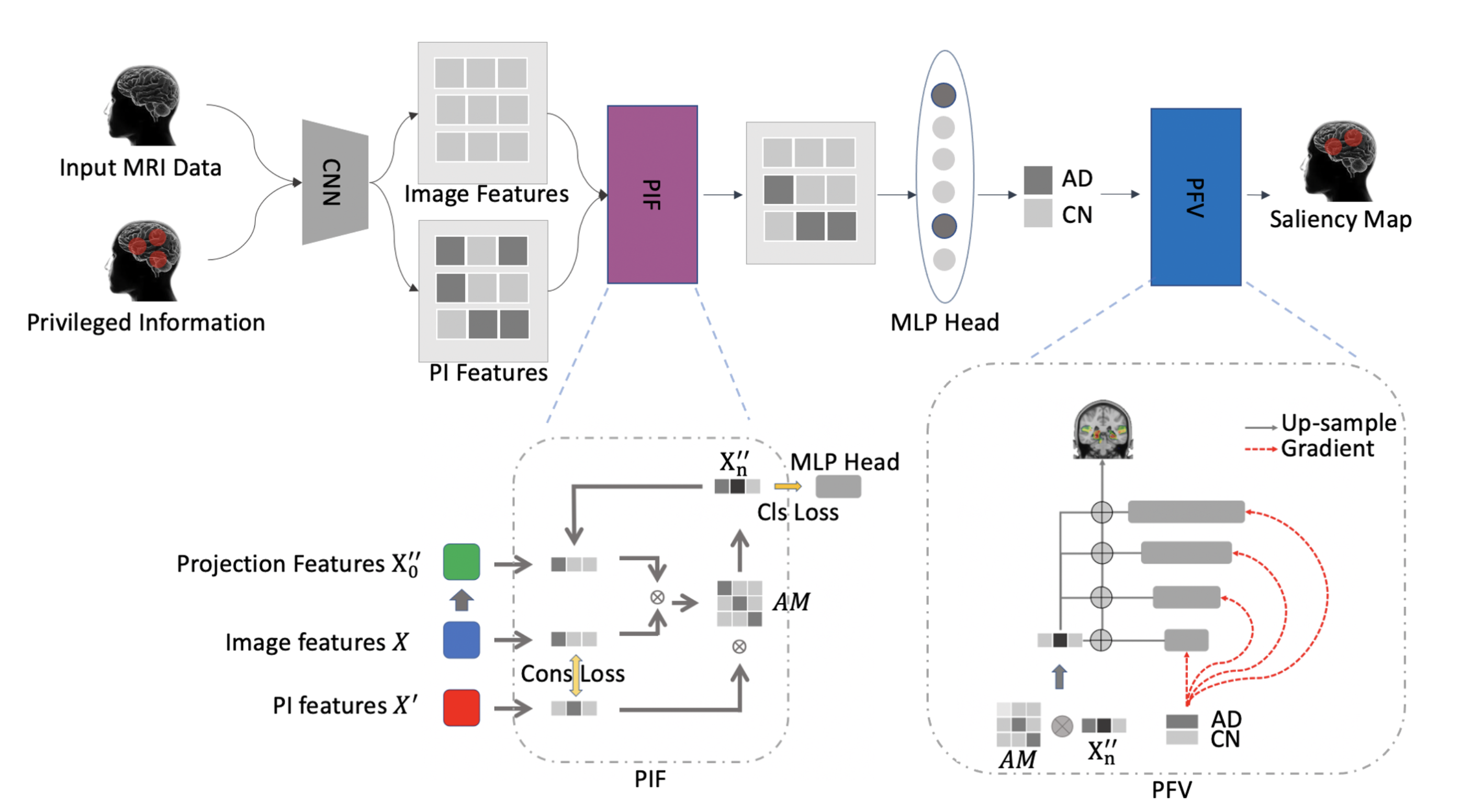
Results
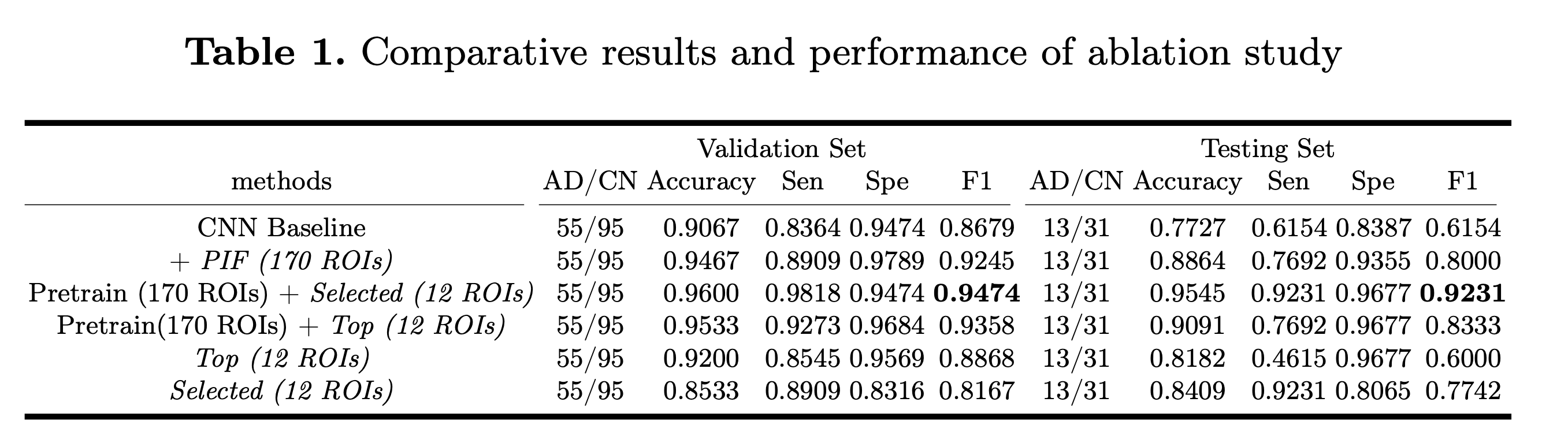
The accuracy and F1 score of our model is 96% (at the third row)
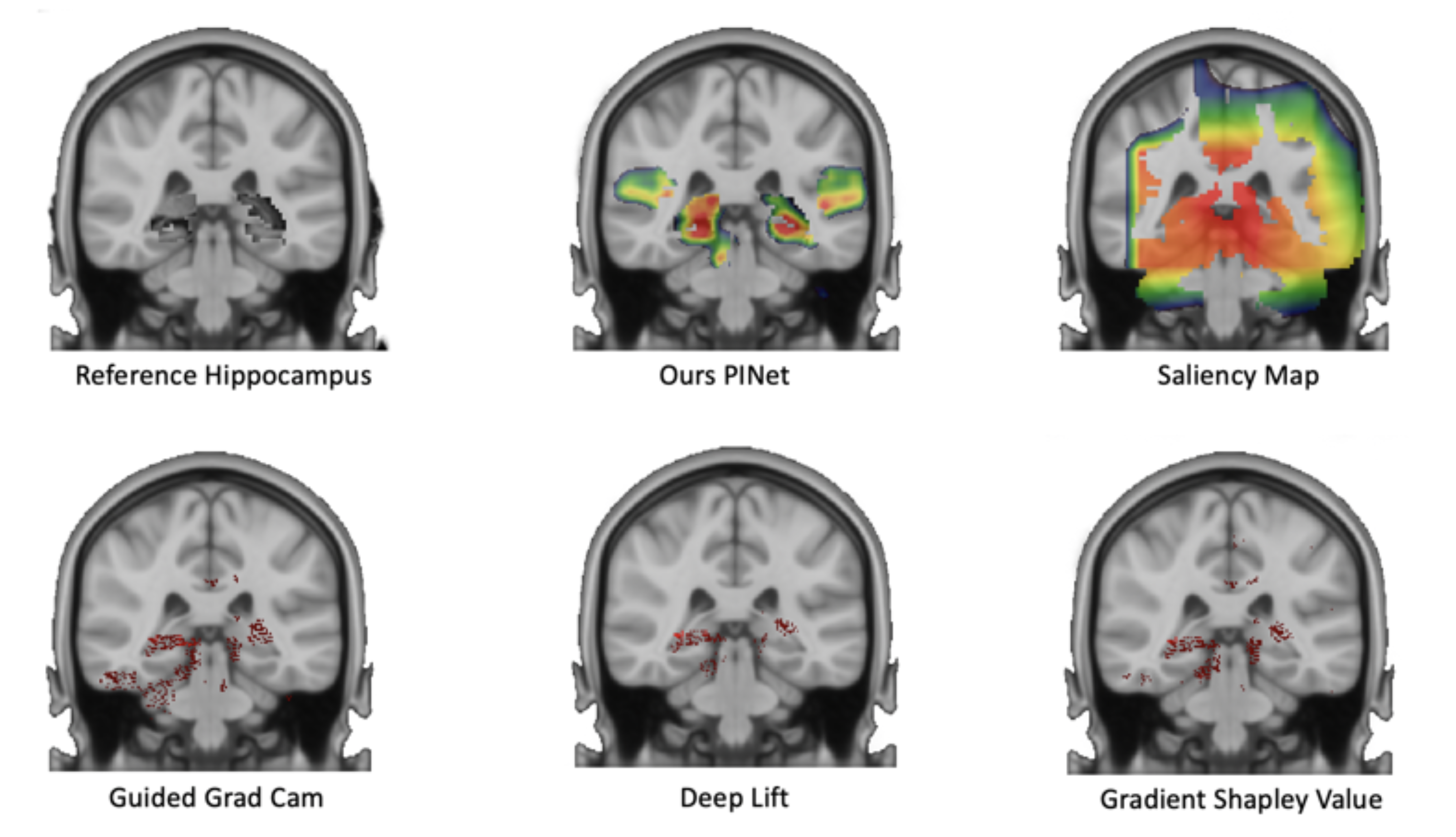
The visualization result of our model.
scPerb: single-cell perturbation via style transfer-based variational autoencoder
Introduction
Hoping to leverage my technical skills to analyze single-cell RNA-seq, I proposed a study to generate single-cell perturbation using generative models. At the current stage, it’s challenging to generate sufficient gene expressions in response to different drugs, doses, and treatments for it’s too labor-intensive and costly: computational tools can fill the gap. Lacking a strong background in bioinformatics, I spent a month reading 50+ papers to familiarize myself with the topic. I first learned by rebuilding one of our top benchmarking models published in Nature Methods, scGen, step by step. scGen is already a top-notch model that’s difficult to improve. However, after rewriting the entire TensorFlow codes into PyTorch-style code and successfully regenerating results, I thought of replacing the fixed linear transformation matrix used in the model to possibly achieve higher potentials. After spending months trying different methods, from multi-layers, multi-encoders, different loss functions like Weighted MSE or ZINB loss and even tinkering with other high-performing models like GANs or VQVAE, I found a rather simple approach: I added style transfer, commonly used for scenarios like transferring the styles of oil paintings to digital images. I innovatively proposed RNA sequences as “styles,” decoupling style-related and content-based features to better “learn” the difference in distribution, mitigating the limitations posed by “vague” images generated by VAEs, and increasing accuracy significantly. Resulted in an $R^2$ value of 0.99.
Abstract
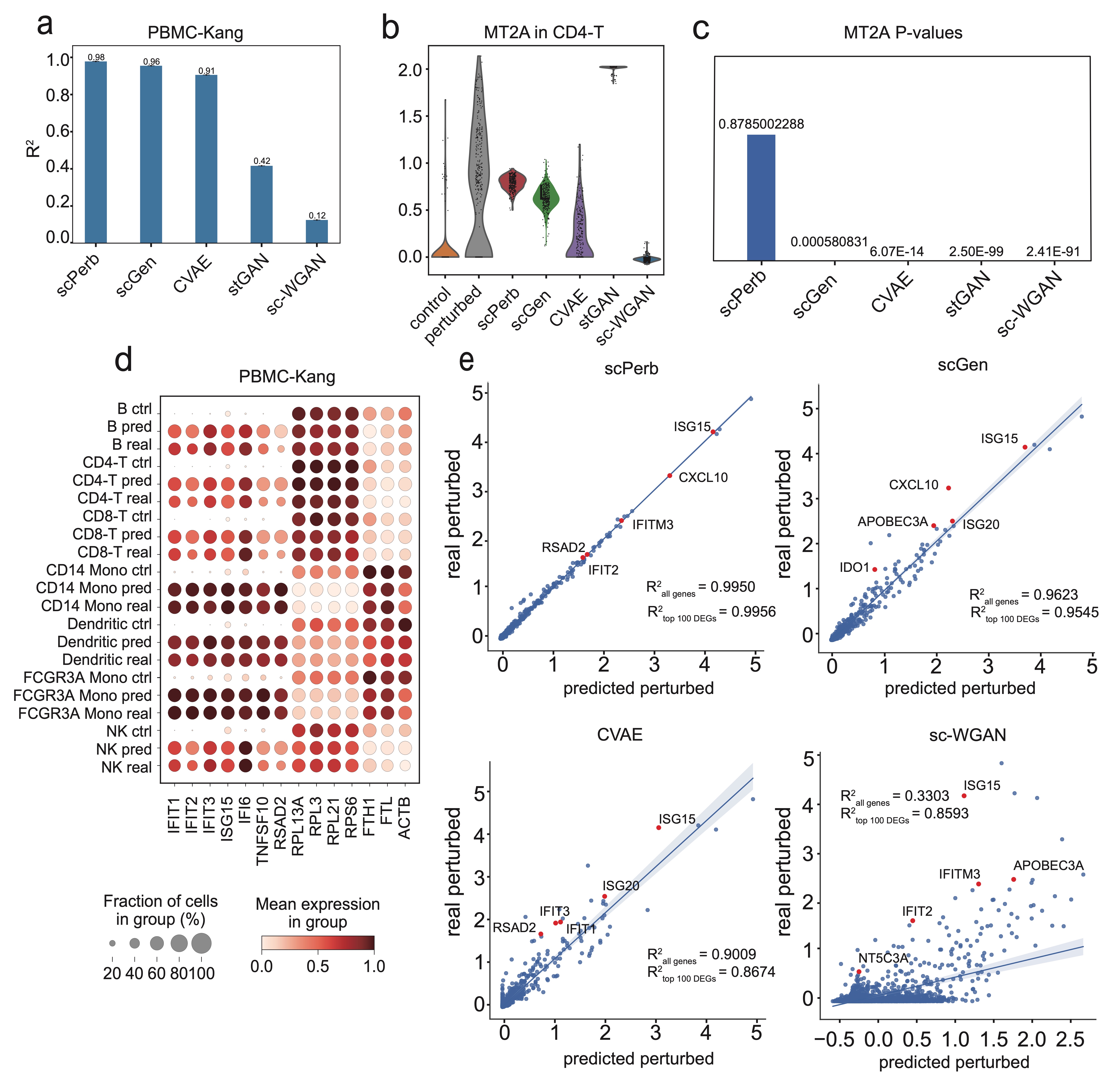
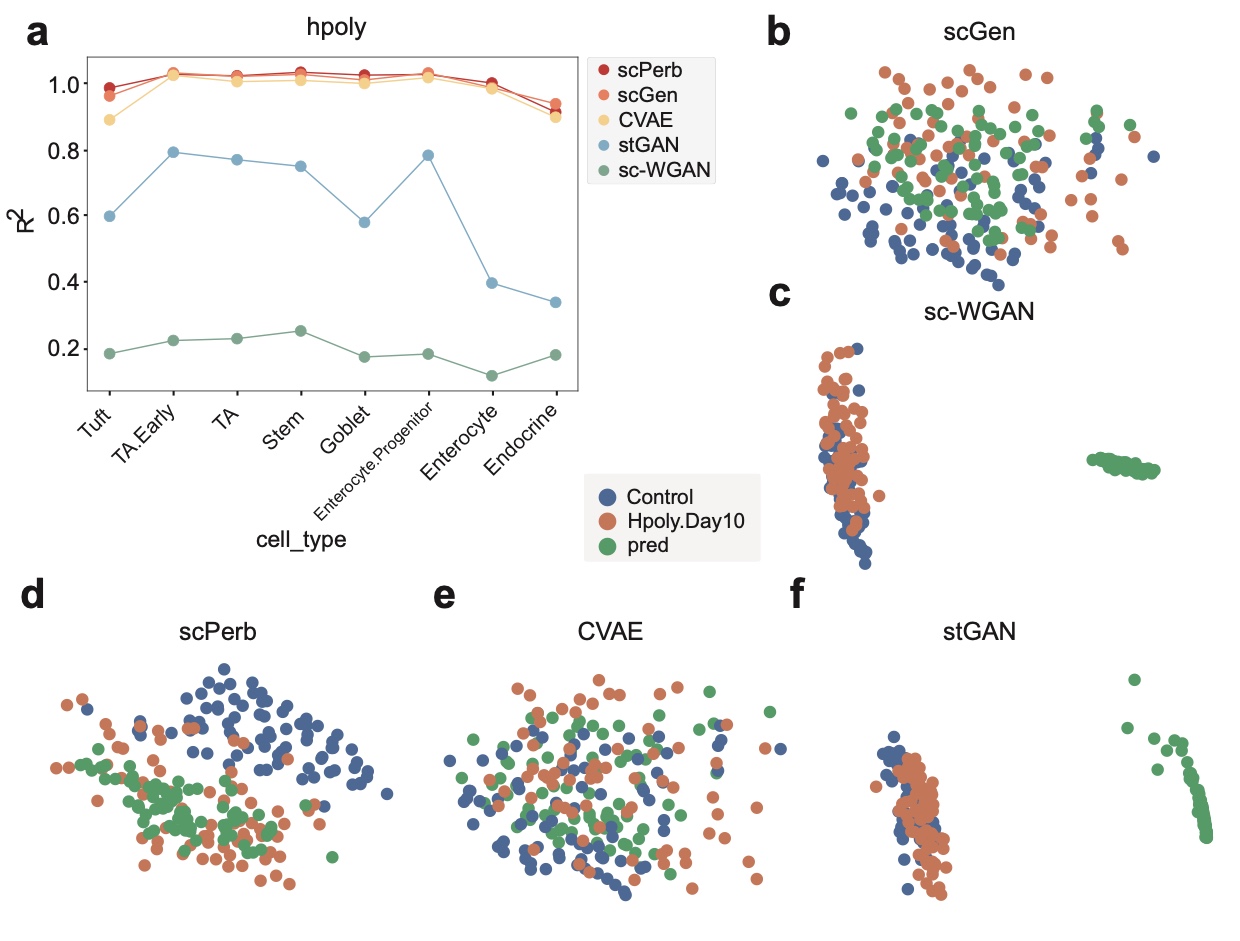
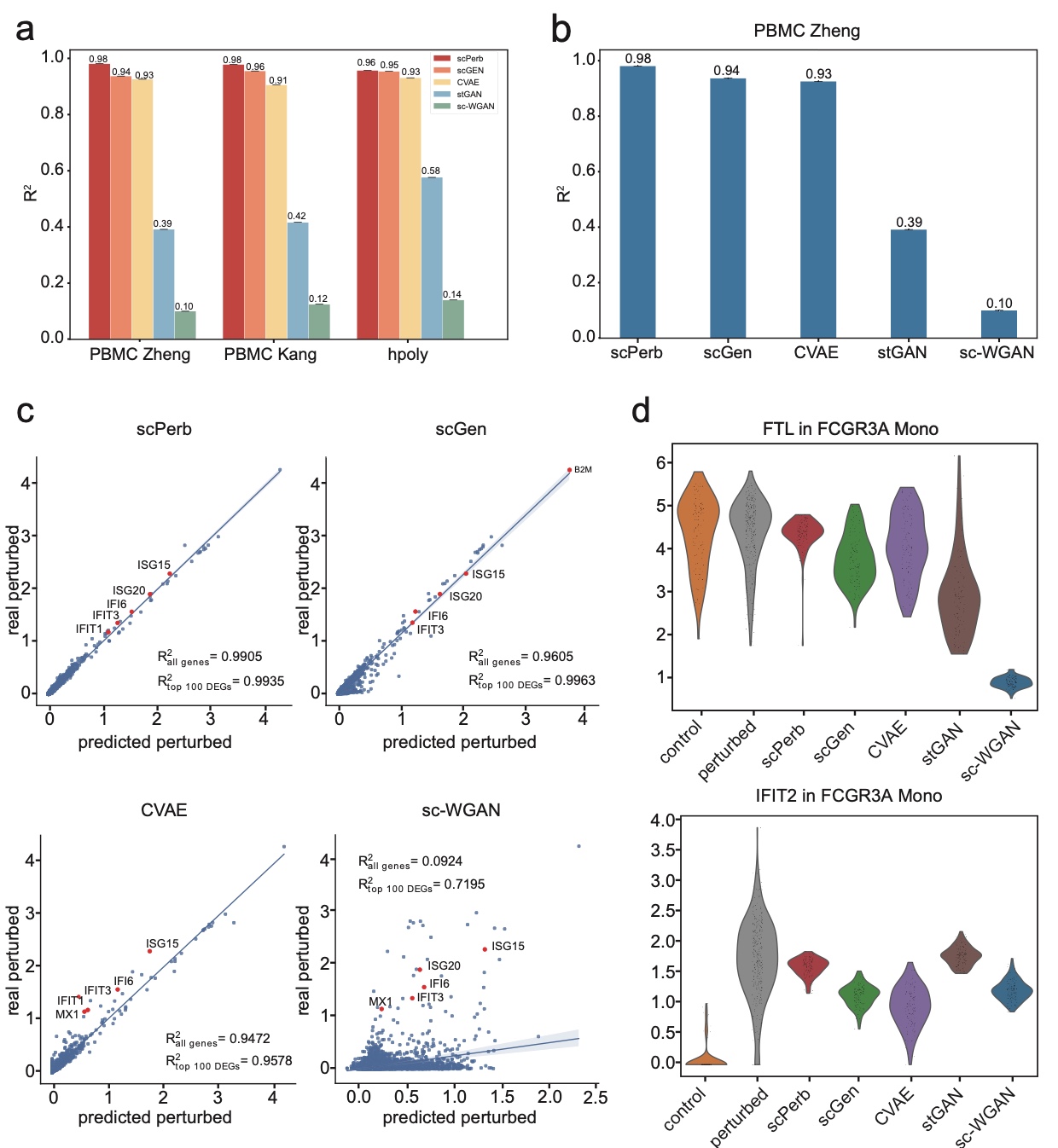
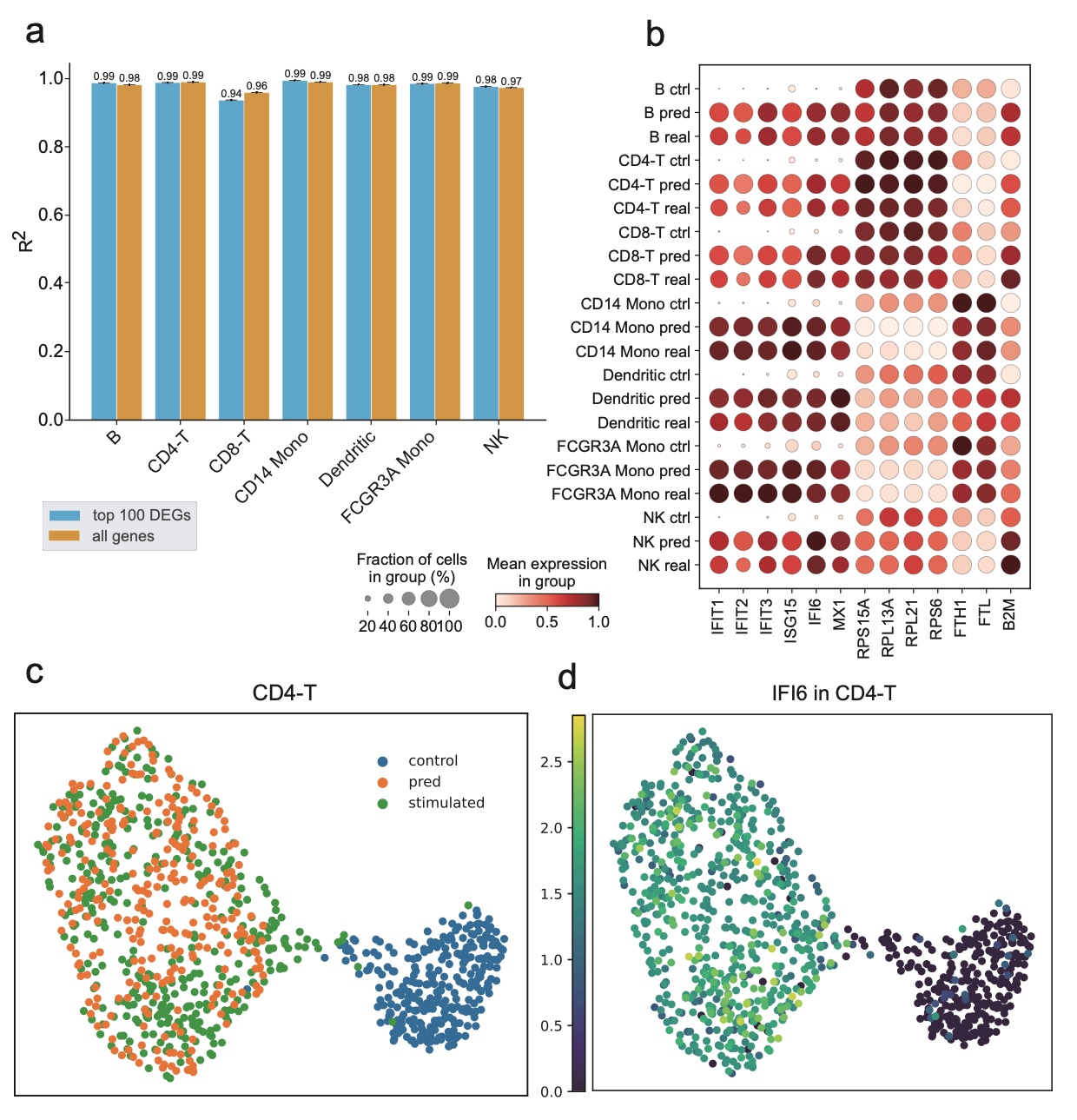
Traditional methods for obtaining cellular responses after perturbation are usually labor-intensive and costly, especially when working with rare cells or under severe experimental conditions. Therefore, accurate prediction of cellular responses to perturbations is of great importance in computational biology. To address this problem, some methodologies have been previously developed, including graph-based approaches, vector arithmetic, and neural networks. However, these methods either mix the perturbation-related variances with the cell-type-specific patterns or implicitly distinguish them within black-box models. In this work, we introduce a novel framework, scPerb, to explicitly extract the perturbation-related variances and transfer them from control data to perturbed data. scPerb adopts the style transfer strategy by incorporating a style encoder into the architecture of a variational for the differences in the latent representations between control cells and perturbed cells, which allows scPerb to accurately predict the gene expression data of perturbed cells. Through the comparisons with existing methods, scPerb presents improved performance and higher accuracy in predicting cellular responses to perturbations. Specifically, scPerb not only outperforms other methods across multiple datasets but also achieves superior $R^2$ values of 0.98, 0.98, and 0.96 on three benchmarking datasets.
Overview
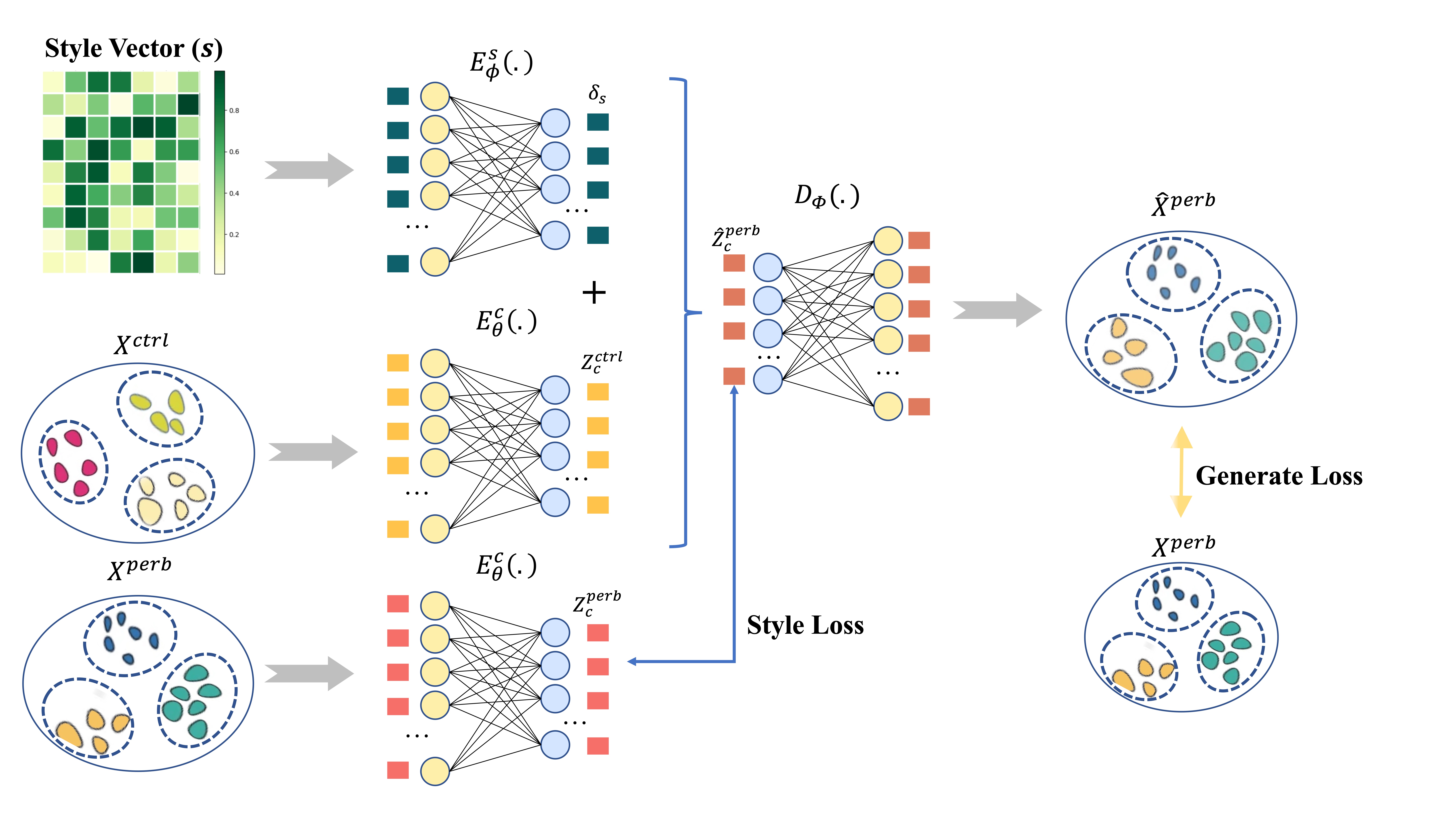
Results